Methods
Following ethics committee approval, all 71 neurologically healthy volunteers provided written informed consent for this prospective study. The cervical spinal cord MRI (C1/2-C7/Th1) was performed in all subjects on a 3T scanner (19-channel head-neck coil) following the protocol outlined in Table 1.
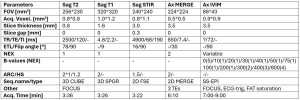
Data processing employed the SpinalCord Toolbox (SCT) [6], DIPY [7], and FSL [8]. Sagittal T2-weighted images were automatically segmented to delineate the spinal cord (sct_deepseg_sc) and identify vertebral body levels (sct_label_vertebrae), with manual annotation of posterior disc positions (sct_label_utils). Axial anatomical images (Ax MERGE) were then segmented into gray matter (GM) and white matter (WM) (sct_deepseg_sc). IVIM data underwent motion artifact correction (sct_dmri_moco), followed by voxel-wise IVIM parameter fitting using DIPY's variable projection method (IvimModelVP). Subsequent steps included segmentation (sct_deepseg_sc), registration of axial anatomical and IVIM images to the PAM50 atlas (sct_register_multimodal), inverse warping of the atlas to individual subject space (sct_warp_template), and extraction of IVIM parameters at C2-C7 (sct_extract_metric) for statistical analysis.
Quality control was implemented throughout the processing pipeline. All SCT commands included the -qc argument, and FSL/DIPY outputs were visually inspected. Processing parameters were adjusted or manual corrections were performed as needed to address errors.
Statistical analyses were conducted at each vertebral level (C2-C7). Welch's t-test, paired t-test, and multivariable linear regression were used to assess the influence of demographic factors. Statistical analyses were performed using R and Statistica 14.